Research Article - (2021) Volume 9, Issue 9
The objective of this study was the isolation, the structure elucidation and the identification of chemical constituents from the n-hexane, the ethyl acetate (EtOAc) and the methanol (MeOH) extracts of a cameroonian propolis sample collected from Ngaoundal. Thirteen secondary metabolites belonging to cycloartane, ursane and oleananetype triterpenes were isolated and their structures were established by chemical tests and detailed spectroscopic analysis as cycloart-23(E)-en-3β,25-diol (1), cycloarta-7(E),23(E)-diene-3β,25-diol (2), cycloarta-23(E),25(26)-diene3β-ol (3), 25-pentoxy cycloart-23(E)-en-3β-ol (4), cycloartenol (5), 3β-hydroxyolean-12-en-18β-28-oic acid (oleanolic acid) (6), 3α-hydroxyoleanolic acid (3-Epi-oleanolic acid (7), 3β-hydroxyurs-12-en-18β-28-oic acid (ursolic acid) (8), 3α-hydroxyursolic acid (3-Epi-ursolic acid) (9), β-amyrin (10), α-amyrin (12), 3-Epi-β-amyrin (11) and 3-Epi-α-amyrin (13). To the best of our knowledge, compounds 2 and 4 are here described for the first time, while compounds 1, 3, 6, 7, 9, 11 and 13 are here reported for the first time in a propolis sample. Their presence in this propolis provides valuable chemotaxonomic information about the plants from which the bees collected resins. The plants origin of this propolis could be Mangifera indica (mango), Orthosiphon stamineus, Boswellia sacra, Boswellia neglecta and Melipona beecheii, Ficus exasperate, Byrosonima fagifolia, Byrosonima crassifolia, Lavandula officinalis, Euphorbia dendroides, Pachysandra terminalis, Melandrium firmum. The n-hexane crude extract (EAP4) was active against S. aureus 209 (MIC: 184 mg/ml) and Candida albicans 62I (MIC: 92 mg/ml). Compounds 1 and 3 exclusively exhibited antimicrobial activity against Salmonella spp (MIC: 0.1-0.2 mg/ml). The MIC values of 1 and 3 were greater than that of the standard drug (Amoxicillin): 0.1-0.2 mg/ml versus 0.4 mg/ml.
Propolis; Isolation; Cycloartane-type triterpenoids; Pentacyclic triterpenoids; Structure elucidation; Chemical structures.
The emergence of new infectious and chronic diseases makes the need for new drugs paramount [1,2]. Although the search for new drugs can begin from different sources, natural products have proven to be one of the richest sources of bioactive ingredients and molecules with privileged scaffolds for the discovery and development of new drugs [3,4].
Propolis is a natural resinous substance collected by bees as Tetragonula sapiens and Apis mellifera species from various plant and tree buds, leaves, exudates and beeswax. Bees use propolis to build their nests, narrow the nest entrances, seal cracks, embalm dead organisms inside the hive and prevent the growth of bacteria and fungi in the nest. Specifically for stingless bees, propolis is also used to construct storage pots for pollen and honey. Propolis is an apicultural product that has been used as an alternative medicine for disease prevention in different parts of the world. The antibiotic properties of propolis provide a healthy hive environment for the honeybee colony.
The chemical composition of propolis depends on the collection site, available plant sources and bee species [5-8]. Propolis contains a variety of compounds, such as polyphenols (flavonoids, phenolic acids, and their esters), terpenoids, steroids, amino acids, waxy acid, and sugars [9-11]. Propolis is well known for its therapeutic properties, including antimicrobial, antitumor, antioxidant and antiparasitic activities, antiviral, anti-inflammatory and anticancer properties [12-15]. A recent study also proved that propolis has antitoxic and antimutagenic activities [16]. All types of bees producing honey can produce propolis, but the generated amount of propolis is different depending on the genus or species of the bees and the flora of the region. The genus that generates a large amount of propolis is Tetragonula, which belongs to a group of stingless bees of the tribe Meliponini [6]. Stingless bees are widespread over tropical and some subtropical regions of the world and they are the major visitors of many flowering plants in the tropics [17,18]. Unlike Apis bees that are bigger and have a functional stinger, Tetragonula bees are smaller and have a non-functional stinger to help defend against nest intruders. They rather use their jaws to bite them [19].
The present work deals with the isolation, the structure elucidation and the identification of the cycloartane-type and pentacyclic terpenoids of Cameroonian propolis.
General experimental procedures
The ESI-TOF- MS spectra (ionization voltage 3kV) in positive mode were measured on a Q-TOF Ultima spectrometer (Waters). Deuterated solvents were used to dissolve the samples for NMR experiments. The NMR spectra (1H, 13C, 1H-1H COSYqf45, HSQC, HMBC and DEPT135) were recorded on four different Brüker spectrometers (500 MHz and 600 MHz) in CDCl3 and DMSO-d6, using TMS as internal standard. Chemical shifts, δ, were expressed in parts per million (ppm) with reference to the solvent residual signals. The coupling constants, J, were expressed in Hertz (Hz). Column chromatography (CC) was performed on silica gel 60 F254 (Ø0.063-0.200 mm, Merck) with step gradients of n-hexane, n-hexane-EtOAc, EtOAc and EtOAc-MeOH as eluents. Crude extracts, fractions and pure compounds were monitored by TLC using Merck pre-coated silica gel sheets (60 F254), and spots were visualized by using UV light ((λmax 254 and 366 nm)) and by spraying with diluted sulfuric acid (50% v/v), followed by heating until colors appeared.
Propolis sample
The Apis melifera propolis samples of Ngaoundal (Mbéré division, Adamawa region) were collected from several hives and scraping from the nests in April 2010 and were supplied by Prof. Tchuenguem Nouhou-Fernand Nestor, Entomologist and Beekeeper at the University of Ngaoundéré, Cameroon.
Extraction and isolation
The triturated raw propolis (875.7 g) was extracted by maceration at room temperature for one week (48 h x 3) with three different solvents (7 L x 3), that is, n-hexane, EtOAc and MeOH, successively. The filtrate was evaporated in a rotatory evaporator and dried by vacuum pump to yield 207.5 g, 115.8 g, 97.6 g of n-hexane, EtOAc and MeOH crude extract s, respectively. A portion of n-hexane (150 g) was subjected to column chromatography (CC) over silica gel, eluted with n-hexane, EtOAc and MeOH in the increasing order of polarities. Two hundred and twentythree sub-fractions of 300 mL each were collected. The solvent was removed under reduced pressure, then, sub-fractions were combined according to their TLC profiles and yielded 34 series or fractions (A1─34). Fraction A9 was purified by successive CC over silica gel and eluted with n-hexane/EtOAc gradient system to give a mixture of 2 and 3. Fractions A11, A12, A13 were purified by re-crystallization to afford 4, 1, and 3, respectively.
A portion of EtOAc (100 g) extract was separated on silica gel CC eluting with n-hexane, n-hexane/EtOAc, EtOAc, EtOAc/MeOH and then MeOH in the order of increasing polarities. Two hundred and sixty-six sub-fractions of 300 mL each were collected and evaporated under reduced pressure with a rotary evaporator (Büchi, 461). The sub-fractions were combined according to their TLC profiles and yielded 21 fractions (B1─21). A white amorphous powder was precipitated in fractions B6 (136 mg) and B9 (92 mg). These series were purified by re-crystallization to afford an inseparable mixture of 10, 12 and 11, 13, respectively. Fraction B15 was washed several times with n-hexane-EtOAc (20%) to give compound 5.
Moreover, MeOH extract (80 g) was also subjected to silica gel column chromatography (Ø 0.063-0.200 mm, 650 g) and eluted with the mixtures of n-hexane-CH2Cl2 and CH2Cl2-MeOH in order of increasing polarity (0-100%) to yield a total of 148 sub-fractions of 250 mL each. These fractions were combined on the basis of TLC analysis into seven major fractions (A1'-A7'). The spots of A1' (19 g) were distinguishable and separable with n-hexane, the mixtures of n-hexane/EtOAc and EtOAc/MeOH as eluent. Fraction A1' (19 g) was subjected to silica gel column chromatographic purification (Ø 0.063-0.200 mm, 500 g) using n-hexane, the mixtures of n-hexane-EtOAc and EtOAc-MeOH with gradient polarity (0-100%) as eluents. Two hundred and three sub-fractions of 100 mL each were collected and combined on the basis of TLC analysis into sixteen series (A1"-A16"). A white amorphous powder was precipitated in fraction A6" (n-hexane-EtOAc: 90/10), whilst yellowish precipitates were formed in fraction A7" the same system. After washing fractions A6" with n-hexane-EtOAc (10%) followed by the filtration, a mixture (151 mg) of 9, 7 was obtained. The same treatment was applied to fraction A7" (n-hexane-EtOAc: 90/10) in which a mixture (143 mg) of 8 and 6 was obtained.
Characterization and identification of the isolated compounds
Fraction A12 (1, 72 mg): white amorphous powder from n-hexane/ EtOAc (85/15) %; ESI+-TOF m/z 463.4831 [M-H2+Na]+(calc.463.3552 for C30H48NaO2), m/z 447.4751 [M-H2O+Na]+(calc. 447.3603 for C30H48NaO); 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction A12 was elucidated as cycloart-23(E)-en- 3β, 25-diol.
Fraction A9 (2, 90 mg): white amorphous powder from n-hexane/EtOAc (90/10) %; ESI+-TOF m/z 463.4871 [M+Na]+(calc.463.3552 for C30H48NaO2), m/z 447.4754 [M- 16+Na]+ (calc. 447.3603 for C30H48NaO); 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction A9 was elucidated as cycloarta- 7(E), 23(E)-diene-3β, 25-diol.
Fraction A13 (3, 195 mg): white amorphous powder from n-hexane/ EtOAc (85/15) %; ESI+-TOF m/z 425.5065 [M+H]+(calc. 425.3783 for C30H49O), m/z 447.4754 [M+Na]+(calc. 447.3783 for C30H48NaO), m/z 871.9832 [2M+Na]+(calc. 871.7308 for C60H96NaO2); 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction A13 was characterized as cycloarta-23(E), 25(26)-diene- 3β-ol.
Fraction A11 (4, 352 mg): white amorphous powder from nhexane/ EtOAc (85/15) %; ESI+-TOF [M-C5H11O]+at m/z 425.5065, base ion peak [M-C5H12O+ Na]+at m/z 447.4754; 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction A11 was characterized as 25-pentoxycycloart-23(E)-en-3β-ol Fraction B15 (5, 135 mg): white amorphous powder from n-hexane/EtOAc (80/20) %, 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction B15 was identified as cycloartenol.
Fraction A7" (143 mg): yellow amorphous powder from nhexane/ EtOAc (90/10) %, 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction A7" was identified as a mixture of 3β-hydroxyolean-12-en-18β-28-oic acid (oleanolic acid; 6), minor isomer and 3β-hydroxyurs-12-en-18β-28-oic acid (ursolic acid: 8), major isomer.
Fraction A6" (151 mg): white amorphous powder from n-hexane/ EtOAc (90/10) %, 1H NMR spectral data (DMSO-d6); 13C NMR (DMSO-d6). Fraction A6" was identified as a mixture of 3α-hydroxyoleanolic acid (3-Epi-oleanolic acid: 7), minor isomer and 3α-hydroxyursolic acid (3-Epi-ursolic acid: 9), major isomer.
Fraction B6 (136 mg): white amorphous powder from n-hexane/ EtOAc (95/5) %, 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction B6 was identified as a mixture of β-amyrin (10), minor isomer and α-amyrin (12), major isomer.
Fraction B9 (92 mg): white amorphous powder from n-hexane/ EtOAc (90/10) %, 1H NMR spectral data (CDCl3); 13C NMR (CDCl3). Fraction B9 was identified as a mixture of 3-Epi-β-amyrin (11), minor isomer and 3-Epi-α-amyrin (13), major isomer.
Antimicrobial assay
Bacterial strains and culture preparations: In this study, Salmonella spp, Escherichia coli and Pseudomonas aeruginosa were isolated from carcass at the Yaoundé slaughterhouse in 2018 according to ISO 6579 (ISO, 2002). The strains, Staphylococcus aureus 209, Escherichia coli WF+ and Candida albicans 62I were obtained from the Bulgarian Type Culture Collection, institute for State Control of Drugs, Sofia. The bacterial strains were maintained on Nutrient Agar slants at 4 °C in the Microbiology Unit of the Laboratory of Food Analysis and Quality Control, Institute of Medical Research and Medicinal Plants Studies (IMPM).
Bacterial suspensions were prepared from loops primarily in buffered peptone water (Oxoid) by incubating at 37 °C overnight. Cultures were then transferred into Nutrient Agar plates (Difco) and incubated at 37°C overnight. For the assay, organisms were subcultured once onto fresh Nutrient Agar and inocula were prepared by transferring colonies to buffered peptone water (9 mL). Following incubation for 2–4 h at 37 °C, bacterial suspensions were adjusted to a turbidity equivalent to a McFarland 0.5 standard by adding sterile buffered peptone water. A sterile swab was used immersed in the bacterial suspension and then pressed onto the wall of the tube to remove excess inocula. Finally the swab was streaked over the entire surface of the Nutrient Agar plates.
Agar plate diffusion assay using analytical paper discs
Preparation of stock solution for the test dilution: Each dried product (1 mg) was weighed and dissolved into 1 mL of 10% DMSO following the method of Kar in a test tube [38]. Three crude extracts of propolis from Ngaoundal, and two pure compounds isolated from n-hexane crude extract (EAP4) were selected for the experiment.
Impregnation of filter-paper discs: Previously analytical paper discs (Ø.12.7mm, Schleider & Schuel, USA) were heat-sterilized at 160°C for 1 h in a hot oven. The discs were then soaked overnight in the stock solution. The solvent was later allowed to evaporate from the discs at 50 °C in a safety cabinet. For each experiment, control disc with pure solvent was used as blind control. The paper discs were applied to the agar plates using a sterilized forceps. To avoid overlapping of the zones of inhibition and possible error in measurement, discs were distributed 24 mm from each other and from the edge of the plate. After 24 h of incubation at 37°C, diameters of zones of growth inhibition were measured in millimeter as described by CLSI [39]. The experiments were carried out in duplicates.
The extracts of propolis sample were submitted to repeated column chromatography to afford two new cycloartane-type triterpenes, cycloarta-7(E),23(E)-diene-3β,25-diol (2) and 25-pentoxycycloart- 23(E)-en-3β-ol (4), together with eleven known secondary metabolites (Figure 1). These known compounds include cycloart-23(E)-en-3β, 25-diol (1) [22], cycloarta-23(E),25(26)- diene-3β-ol (3) [23], cycloartenol (5) [20, 21], 3β-hydroxyolean-12- en-18βH-28-oic acid (oleanolic acid) (6) [25], 3α-hydroxyoleanolic acid (3-Epi-oleanolic acid (7) [23], 3β-hydroxyurs-12-en-18β-28-oic acid (ursolic acid) (8) [25], 3α-hydroxyursolic acid (3-Epi-ursolic acid) (9) [23], β-amyrin (10) [28-30], 3-Epi-β-amyrin (11) [26, 27], α-amyrin (12) [28-30] and 3-Epi-α-amyrin (13) [26, 27].
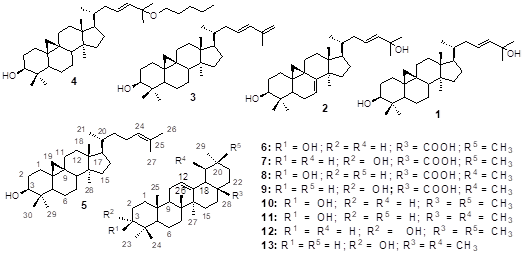
Figure 1: Structure of the isolated compounds
Compounds 1-13 were isolated from the fractions Hex/EtOAc (80-85/10-20%) as white amorphous powder and gave positive test with Liebermann-Burchard reagent, showing that they are triterpenoids. The ESI-TOF MS positive mode of 1 exhibited a pseudomolecular ion peak [M-H2+Na]+ at m/z 463.4831 (calc. 463.3552 for C30H48NaO2), a base ion peak [M-H2O+Na]+ at m/z 447.4751 (calc. 447.3603 for C30H48NaO). Its molecular formula was deduced as C30H50O2, indicating six degrees of unsaturation. The molecular formula of 2 was determined as C30H48O2 according to the positive ESI-TOF MS from the [M+Na]+ and [M-16+Na]+ signals at m/z 463.4871 (calc.463.3552 for C30H48NaO2) and at m/z 447.4754 (calc. 447.3603 for C30H48NaO), respectively, indicating seven degrees of unsaturation. The ESI-TOF MS in positive mode of 3 showed the ion peaks [M+H]+ at m/z 425.5065 (calc. 425.3783 for C30H49O), the base peak ion [M+Na]+ at m/z 447.4754 (calc. 447.3783 for C30H48NaO) and [2M+Na]+ at m/z 871.9832 (calc. 871.7308 for C60H96NaO2), which close examination with the 13C-NMR spectrum led to the molecular formula C30H48O with seven degrees of unsaturation. The molecular formula of 4 was determined as C35H60O2 according to the ESI+- TOF MS from the ion signals: [M-C5H11O]+ at m/z 425.5065 (calc. 425.3783 for C30H49O) and [M-C5H12O+Na]+ at m/z 447.4754 (calc. 447.3603 for C30H48NaO), indicating six degrees of unsaturation. The ESI-TOF MS positive mode of 5 indicated the base peak ion [M+Na]+ at m/z 449.3735 (calc. 447.3759 for C30H48NaO), which close examination with the 13C-NMR spectrum led to the molecular formula C30H50O with six degrees of unsaturation, indicating two protons more than that of 2 and 3. Its molecular formula was identical to that of 1 and 4. The NMR spectra of compounds 1-5 was compatible with that of a tetracyclic dammarane ring with the same patterns of HSQC edited and HMBC spectra except for the difference in the chemical shifts around C-3, C-20 and C-25. The compounds with OH group at C-3 (1- 5) expressed the δH-3 at about 3.12 and the δC-3 at 78.8. The 1H NMR spectral data (Table 2) for 1-5 (except H-26 of 3) showed two methyl groups as singlets at δH around 1.13-1.40 and 1.13- 1.76 for H-26 and H-27, respectively. The doublet (J23-24 = 15.0 Hz) and the doublet of triplet (J23-24 = 15.0, J22-23 = 7.0 Hz), (J23-24 = 11.0, J22-23 = 7.8 Hz) of compounds 1, 2, 4 and 3 due to one proton each at δH 5.42-5.54 and 5.3-6.03 were attributed to the olefinic protons at positions C-24 and C-23, respectively. The coupling constant value indicated E-configuration of double bond. The signals of two methyl groups (C-26 and C-27) bonding to C-25 and olefinic carbons (C-23 and C-24) were assigned through the HMBC correlations of H-26 and H-27 with C-24 and C-25, of H-23 with C-25, as well as with the correlations of H-22 with C-23 and C-24 (Figure 3). Another differences among these compounds were the absence of an OH group at C-20 for all of them and the presence of an OH group at C-25 (δC 70.77-70.80) for 1, 2, 4 and δC 129.7, 130.9 for 3 and 5, respectively. From the HSQC edited and HMBC spectra of compound 2, there were two additional signals of sp2 hybridized carbons at δC 115.1 and 148.0 attributed to C-7 and C-8 positions compared to that of 1 and 4 having the same oxygen atom at C-25 (δC 70.7 and 70.8, respectively) and the same sp2 hybridized carbons, C-23 and C-24 (δC 125.6, 139.3-139.4, respectively). Adversely, the δC of C-26 and C-27 of 5 (δC 17.7, 25.7, respectively), then δC of C-27of 3 (δC 18.8) were shifted upfield compared to δC around 29.9 of 1 and 4. The HMBC spectra of these compounds (Figure 3) showed the normal correlations of H-21 with C-17 and C-20, moreover, H-27 of 3 showed cross-peak correlations with C-26 (δC 114.0) and C-24 (δC 139.3). In addition, the same 1H-NMR spectral data (Table 2) of compounds 1-5 showed a set of AB doublets at δH 0.18-0.33 and 0.39-0.55 (each 1H, d, J = 4.1-5.8 Hz) characteristic of a cyclopropane moiety methylene protons Hα- 19 and Hβ-19, respectively; four other tertiary methyl groups at δH 0.78-1.16, 1.13-1.19, 0.69-0.89, 0.78-1.22 and 0.62-0.82; one secondary methyl group at δH 0.67-0.92 (1H, d, J = 6.6 Hz). The same 13C-NMR (Table 1), HSQC and DEPT135 spectra of 1-5 indicated the four other tertiary methyl groups at δC 18.0-18.1 (C-18), 19.3 (C-28), 25.4 (C-29) and 14.0 (C-30); one secondary methyl group at δC 18.2-18.6 (C-21) and a C-9/C-19 cyclopropyl methylene at δC 29.9-30.0 (C-19).
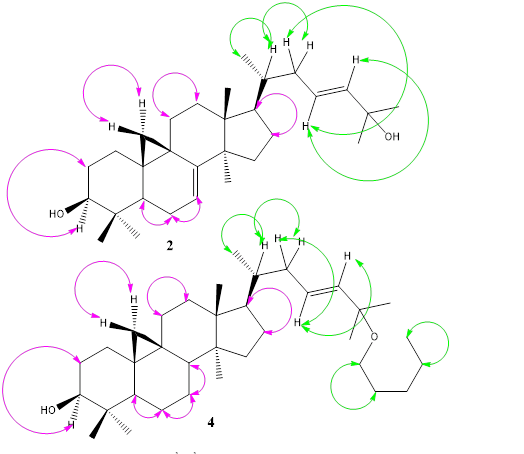
Figure 2: Selected 1H-1HCOSY correlations of compounds 2 and 4
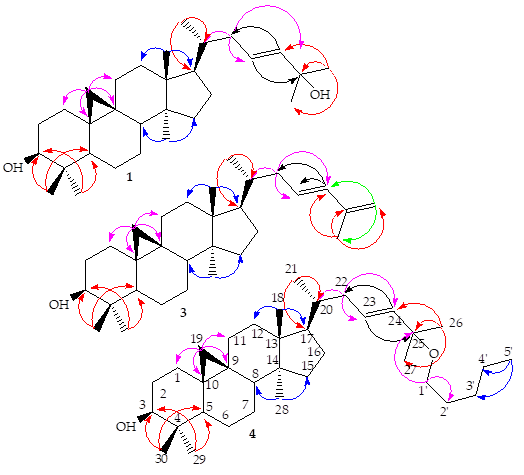
Figure 3: Selected HMBC correlations of compounds 1, 3 and 4
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 32 | 32 | 32 | 32 | 32 | 39.4 | 38.2 | 38.6 | 38.2 | 38.6 | 38.2 | 38.7 | 38.2 |
| 2 | 30 | 30.4 | 30.4 | 30.1 | 30.4 | 27.2 | 27.5 | 26.7 | 27.5 | 27.2 | 27.5 | 27.4 | 27.5 |
| 3 | 78.8 | 78.8 | 78.8 | 78.8 | 78.9 | 78 | 76.8 | 78.7 | 76.8 | 79 | 76.2 | 79.1 | 76.2 |
| 4 | 40.5 | 40.5 | 40.5 | 40.5 | 40.5 | 39.7 | 38.8 | 39.1 | 38.8 | 38.8 | 38.8 | 38.8 | 38.8 |
| 5 | 52 | 52 | 52.2 | 52 | 52.3 | 55.7 | 54.8 | 55.2 | 54.8 | 55.2 | 54.8 | 55.3 | 54.8 |
| 6 | 21.1 | 21.1 | 21.1 | 22.7 | 21.1 | 18.7 | 18 | 18.2 | 18 | 18.4 | 18 | 18.4 | 18 |
| 7 | 26.1 | 115 | 26.1 | 26.5 | 26.5 | 33.2 | 33.3 | 32.9 | 33.3 | 32.6 | 33.3 | 32.8 | 33.3 |
| 8 | 48 | 148 | 48 | 48 | 48.5 | 46.6 | 40.1 | 39.4 | 40.1 | 39.8 | 40.1 | 40.8 | 40.1 |
| 9 | 20 | 20 | 20 | 20 | 20 | 48 | 47.1 | 47.5 | 47 | 47.6 | 47.1 | 49.7 | 47 |
| 10 | 26 | 26 | 26 | 26 | 26 | 37.3 | 36.6 | 36.8 | 36.6 | 36.9 | 36.6 | 36.9 | 36.6 |
| 11 | 26.4 | 26.4 | 26.4 | 27.1 | 26.5 | 23.7 | 23.8 | 23.2 | 23.8 | 23.5 | 23.8 | 23.3 | 23.8 |
| 12 | 32.8 | 32.8 | 32.8 | 32.8 | 32.9 | 123 | 121.5 | 125 | 124.6 | 121.7 | 121.5 | 121.7 | 124.6 |
| 13 | 45.3 | 45.3 | 45.3 | 45.3 | 45.3 | 145 | 143.8 | 138 | 138.2 | 145.2 | 143.8 | 145.2 | 138.2 |
| 14 | 48.8 | 48.8 | 48.8 | 48.8 | 48.8 | 42.1 | 41.6 | 42 | 41.3 | 41.8 | 41.6 | 42.1 | 41.3 |
| 15 | 35.6 | 35.6 | 35.6 | 36.4 | 35.6 | 26.9 | 27.2 | 30 | 27.2 | 26.1 | 27.2 | 28.4 | 27.2 |
| 16 | 28.1 | 28.1 | 28.2 | 29.4 | 28.1 | 23.6 | 22.8 | 24.1 | 27 | 26.9 | 22.8 | 26.6 | 27 |
| 17 | 47.1 | 47.1 | 47.1 | 47.1 | 47.1 | 46.6 | 46.8 | 47.7 | 46.8 | 32.5 | 46.8 | 33.8 | 46.8 |
| 18 | 18.1 | 18.1 | 18.1 | 18.1 | 18 | 42.1 | 40.8 | 52.7 | 52.3 | 47.2 | 40.8 | 59.1 | 52.3 |
| 19 | 29.9 | 30 | 30 | 30 | 29.9 | 46.6 | 45.6 | 39 | 38.5 | 46.8 | 45.6 | 39.8 | 38.5 |
| 20 | 36.4 | 36.4 | 36.8 | 36.4 | 35.9 | 30.9 | 30.4 | 38.8 | 38.4 | 31.1 | 30.4 | 40.1 | 38.4 |
| 21 | 18.3 | 18.3 | 18.3 | 18.8 | 18.2 | 32.4 | 36.3 | 30.7 | 30.1 | 34.7 | 36.3 | 31.3 | 30.1 |
| 22 | 39 | 39 | 39.7 | 37.1 | 35 | 33.2 | 32.7 | 36.7 | 36.3 | 37.1 | 32.7 | 41.6 | 36.3 |
| 23 | 126 | 126 | 130 | 125.6 | 24.9 | 28.8 | 28.2 | 27.9 | 28.2 | 28.1 | 28.2 | 28.6 | 28.2 |
| 24 | 139 | 139 | 134 | 139.3 | 125.3 | 16.5 | 15.1 | 15.2 | 15.1 | 15.5 | 15.1 | 15.4 | 15.1 |
| 25 | 70.7 | 70.7 | 142 | 70.8 | 130.9 | 15.6 | 15.2 | 15.5 | 15.2 | 15.6 | 15.2 | 15.6 | 15.2 |
| 26 | 25.4 | 27.1 | 114 | 29.9 | 17.7 | 17.4 | 16.9 | 16.7 | 16.9 | 16.8 | 16.8 | 16.8 | 16.9 |
| 27 | 29.9 | 27.1 | 18.8 | 29.9 | 25.7 | 26.2 | 25.6 | 23.4 | 23.3 | 26 | 25.6 | 23.1 | 23.3 |
| 28 | 19.3 | 19.3 | 19.3 | 19.3 | 19.3 | 180 | 178.6 | 181 | 178.3 | 28.4 | 28.4 | 28.4 | 28.4 |
| 29 | 30.4 | 25.4 | 25.4 | 25.4 | 25.4 | 32.9 | 32.8 | 16.9 | 17 | 33.3 | 32.8 | 17.4 | 17 |
| 30 | 14 | 14 | 14 | 14 | 14 | 26.1 | 23.2 | 21 | 21.1 | 23.7 | 23.2 | 21.3 | 21.1 |
| 1′ | - | - | - | 67.5 | - | - | - | - | - | ||||
| 2′ | - | - | - | 29.7 | - | - | - | - | - | ||||
| 3′ | - | - | - | 29.4 | - | - | - | - | - | ||||
| 4′ | - | - | - | 22.7 | - | - | - | - | - | ||||
| 5′ | - | - | - | 14.2 | - | - | - | - | - |
Table 1:.13C (500 MHz) NMR data for compounds 1-9 in CDCl3 (δC in ppm)
The 1H-1H COSY experiments (Figure 2) of 1-5 showed crosspeak correlations between H-23/H-24, H-23/H-22, H-20/H-21, H-20/H-22, H-3 /H-2, H-5/H-6, H-6/H-7, H-12 /H-11, H-19β/ H-19α, and H-1'/H-2', H-5'/H-4' (compound 4).
All the above evidences confirmed that compounds 1-5 were all 9,19-cycloartane triterpenes and not dammarane-type triterpenes. The remaining spectral data showed some similarities with those reported data on cycloartane triterpenoidal skeleton [20, 21]. Thus, compounds 1 was identified as cycloart-23(E)-ene-3β, 25- diol [22], compound 2 as cycloarta-7(E), 23(E)-diene-3β,25-diol, compound 3 as cycloarta-23(E), 25(26)-diene-3β-ol [23] and compound 4 as 25-pentoxycycloart-23(E)-en-3β-ol. Finally, the comparison of NMR data of compound 5 to those reported in the literature [20, 21] made its identification as cycloartenol.
To the best of our knowledge, compounds 2 and 4 are here described for the first time. Their presence in this propolis provides valuable chemotaxonomic information about the plants from which the bees collected their resins.
The 1HNMR spectral data (Figure S12, Table 2) of 7 and 9 displayed a broad singlet at δH 11.97 attributed to two symmetrical protons of two hydroxyl groups linked each to one acyl of the carboxylic acid; two downfield chemical shift values at δH 5.16 and 5.13 (both triplet) assigned to two olefinic protons (2 H-12) bonded to 2 C-12 position, while the doublet of doublet of two oxymethine protons bound to 2 C-3 position resonated at δH 3.00 for the two isomers found in ring C and A of the oleanane and ursane triterpenoid series. This spectrum also revealed a set of broad double doublet at δH 2.75 and 2.72 which were assigned to H-18β of oleanane skeleton, and one doublet at δH 2.11 corresponding to H-18β of the ursane skeleton, and then the presence of the angular methyl proton signals at δH 1.04 (3H, s, H-23), δH 0.82 (3H, s, H-24), δH 0.68 (3H, s, H-25), δH 0.85 (3H, s, H-26), δH 1.04 (3H, s, H-27), δH 0.90 (3H, d, H-29), δH 0.87 (3H, d, H-30) for the major isomer, and δH 1.04 (3H, s, H-23), δH 0.82 (3H, s, H-24), δH 0.75 (3H, s, H-25), δH 0.85 (3H, s, H-26), δH 1.09 (3H, s, H-27), δH 0.87 (3H, s, H-29), δH 0.81 (3H, s, H-30) for the minor isomer. The 13C NMR (Figure S13, Table 2, DMSO-d6, 67.5 MHz) and DEPT 135 spectra of 7 and 9 exhibited two quaternary carbon signals at δC 178.6 and 178.3 (2C-28) assigned to two carboxyl groups of the carboxylic acids; four olefinic carbon signals at δC 121.5 and 124.6 (2C-12), δC 138.2 and 143.8 (2C-13); two symmetrical oxymethine carbon signals at δC 76.8 (2C-3) and fourteen angular methyl carbon signals appearing at δC 28.2 (C-23), 15.1 (C-24), 15.2 (C-25), 16.9 (C-26), 23.3 (C-27), 17.0 (C-29), 21.1 (C-30) for the major isomer, and at δC 28.2 (C-23), 15.1 (C-24), 15.2 (C-25), 16.9 (C-26), 25.6 (C-27), 32.8 (C-29), 23.2 (C-30) for the minor isomer. The HMBC spectrum clearly indicated the correlations of 2 H-16, 1 H-18, 2 H-22 with the carbonyl carbon signals at δC 178.6 and 178.3. The absence of the methyl carbon signals at δC 28.4 and 28.1 [22] proved that the angular methyl groups at C-28 were oxidized to the carboxylic acids. The 1HNMR spectral data (Table 2) of 7 and 9 were consistent with those of 3α-hydroxyoleanolic acid (3-Epi-oleanolic acid) and 3α-hydroxyursolic acid (3-Epi-ursolic acid) previously synthesized by Wen et al. (2008) [24]. This author has not reported on the 13C NMR spectral data. The 1H NMR, 13C NMR and DEPT 135 data of 7 and 9 were also closely similar to those isolated and reported by Hatem and Najah (2016) [25]. The 2D-NMR (HSQC, HMBC and 1H-1H COSY) experiments of compounds 7 and 9 presented some similarities with those of 6 and 8 except for the difference in the chemical shifts around C-3 and C-28. Compounds 6 and 8 with the OH group at C-3 expressed the δH-3 about 3.44 and the δC-3 78.0, 78.7 instead of δH-3 3.00 and δC-3 76.8; furthermore, δC-28 180.2, 180.6 for 6 and 8 shifted to the downfield compared to that of 7 and 9. We also observed that the chemical shift values around C-3 at δC-3 76.2 for 11 and 13 shifted to downfield compared to compounds 7, 9 (both δC-3 76.8). They expressed slight differences around δH-3 about 3.17 for compound 11 and 3.44 for 13 which were identical to those of 6 and 8. The compounds with a COOH group at C-28 (6-9) expressed the δC-28 about 178.3-180.6, whereas compounds 10-13 didn’t show these chemical shift values at the same position, but presented δC-28 28.4 each. From the HSQC edited and HMBC spectra, the oxymethine tertiary carbon signal of C-3 at δC-3 79.0- 79.1 of 10 and 12 shifted downfield compared to δC-3 78.0-78.7 of 6 and 8. Therefore, compounds 7 and 9 were identified as a mixture of 3α-hydroxyolean-12-en-18β-28-oic acid and 3α-hydroxyurs- 12-en-18β-28-oic acid; 6 and 8 as a mixture of 3β-hydroxyolean- 12-en-18β-28-oic acid (oleanolic acid) and 3β-hydroxyurs-12-en- 18βH-28-oic acid (ursolic acid) previously isolated by Hossain and Ismail (2013) [26] from the leaves of Orthosiphon stamineus; 11, 13 as 3-Epi-β-amyrin and 3-Epi-α-amyrin [27,28] previously isolated from the gum resins of Boswellia neglecta and Boswellia sacra . Finally, compounds 10 and 12 identified as β-amyrin and α-amyrin previously identified by GC-MS analysis of the chloroform-methanol propolis extract from Melipona beecheii and isolated [29-32]. To the best of our knowledge compounds 1, 3, 6, 7, 9, 11 and 13 are here reported for the first time from propolis source. They are new propolis constituents and their presence in this propolis provides valuable chemotaxonomic information about the plants from which the bees collected their resins.
| H/C | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| δH multiplicity (J in Hz) | |||||||||||||
| 1 | 1.61 m, 1.24 m | 1.61 m, 1.24 m | 1.25 m, 1.51 m | 1.62 m, 1.24 m | 1.62 m, 1.24 m | 1.02 m, 157 m | 1.00 m, 1.58 m | 1.00 m, 1.60 m | 1.73 m, 1.03 m | 1.73 m, 1.03 m | 2.01-2.23 m | 1.00 m, 1.60 m | |
| 2 | 1.75 m, 1.59 m | 1.75 m, 1.59 m | 1.32 m, 1.75 m | 1.75 m, 1.62 m | 1.75 m, 1.52 m | 1.82 m | 1.81 m | 1.82 m | 1.67 m, 1.61 m | 1.67 m, 1.61 m | 1.82 m | 1.82 m | |
| 3 | 3.12 dd | 3.12 dd | 3.30 dd | 3.22 dd | 3.29 m | 3.44 dd | 3.60 br s | 3.44 dd | 3.00 dd | 3.27 dd (11.1, 4.3) | 3.17 br s | 3.21 dd (11.5, 4.5) | 3.39 br s |
| (6.4, 9.2) | (6.4, 9.2) | ( 12.5, 4.5) | ( 11.1, 4.8) | - | - | - | - | - | - | - | - | ||
| 4 | - | - | - | - | - | 0.88 dd | 0.88 dd | 0.90 m | 0.80 dd (14.2, 4.2) | 0.80 dd (14.2, 4.2) | 0.93 s | 0.90 m | |
| 5 | 1.31 m | 1.31 m | 1.32 dd (6.0, 2.5) | 1.37 m | 1.33 m | 1.58 m, 1.39 m | 1.58 m, 1.39 m | 1.35 m | 1.59 m, 1.44 m | 1.59 m, 1.44 m | 1.35 m | 1.35 m | |
| 6 | 1.42 m, 0.85m | 1.42 m, 0.85m | 0.80 m, 1.60 m | 1.49 m, 0.78 m | 1.49 m, 0.78 m | 1.53 m, 1.36 m | 1.59 m, 1.39 m | 1.55 m | 1.58 m, 1.38 m | 1.58 m, 1.38 m | 1.55 m | ||
| 7 | 1.31 m , 1.15 m | 6.06 d (6.4) | 1.10 m, 2.01 m | 1.49 m, 0.98 m | 1.31 m, 1.12 m | - | - | - | - | - | - | - | - |
| 8 | 1.51-162 m | - | 1.54 dd ( 3.5, 2.5) | 1.66-1.82m | 1.55–1.62 m | 1.71 t | 1.59 t | 1.62 t | 1.59 m | 1.59 m | 2.04 t | 1.62 t | |
| 9 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 10 | - | - | - | - | - | 1.96 m | 1.96 m | 1.95 m | 1.95 m, 1.91 m | 1.95 m, 1.91 m | 1.95 m | 1.95 m | |
| 11 | 2.03 m, 1.10 m | 2.03 m, 1.10 m | 1.03 m, 2.01 m | 2.12 m | 2.03 m, 1.16 m | 5.49 s | 5.49 br s | 5.49 s | 5.16 t | 5.19 t (3.2) | 5.08 br s | 5.16 t (3.2) | 5.12 br s |
| 12 | 1.51–1.62 m | 1.51–1.62 m | 1.62 m | 1.75-1.87 m | 1.61–1.62 m | - | - | - | - | - | - | - | |
| 13 | - | - | - | - | - | - | - | - | - | - | - | - | |
| 14 | - | - | - | - | - | 1.22 m, 2.19 m | 1.22 m, 2.33 m | 1.20 m | 1.81 m, 1.01 m | 1.81 m, 1.01 m | 1.85 m | 1.20 m | |
| 15 | 1.25–1.35 m | 1.25–1.35 m | 1.30 m | 1.28–1.32 m | 1.28–1.32 m | 2.35 br t | 2.35 brt | ||||||
| 16 | 1.82 m, 1.27 m | 1.82 m, 1.27 m | 1.30 m, 1.92 m | 1.87 m, 1.37 m | 1.90 m, 1.27 m | 2.12 t | 2.14 m, 201 m | 2.06 m, 2.50 m | 2.05 m, 0.86 m | 2.05 m, 0.86 m | 1.47 m, 2.24 m | 2.06 m, 2.50 m | |
| 17 | 1.82 m, 1.28 m | 1.82 m, 1.28 m | 1.59 m | 1.58–1.61 m | 1.58–1.61 m | - | - | - | - | - | - | - | |
| 18 | 0.78 s | 0.78 s | 0.89 s | 1.16 s | 0.97 s | 3.30 dd | 2.75 d (15.5) | 2.63 d | 2.11 d (15.5) | 1.99 m | 1.99 m | 1.60 d (11.5) | 2.63 d |
| 19 | 0.18 d (5.8) | 0.18 d (5.8) | 0.25 d (4.0) | 0.27 d (4.0) | 0.32 d (4.1) | 2.72 d (15.5) | 1.71 m, 1.06 m | 1.71 m, 1.06 m | 1.80 m | 1.50 m | |||
| 0.39 d (5.8) | 0.39 d (5.8) | 0.49 d ( 4.0) | 0.51 d ( 4.0) | 0.54 d (4.1) | 1.83 m, 1.32 m | 1.49 dd | 1.50 m | - | - | - | 1.05 m | ||
| 20 | 1.28–1.36 m | 1.28–1.36 m | 1.49 m, 2.19 m | 1.28–1.32 m | 1.28–1.32 m | - | 1.05 m | 1.05 m | 1.42 d (12.2) | 1.42 d (12.2) | 1.51 m | 1.35 m, 1.55 m | |
| 21 | 0.67 d (6.6) | 0.67 d (6.6) | 0.76 d (6.6) | 0.67 d (6.6) | 0.91 d (6.6) | 1.46 m, 1.23 m | 1.40 m, 1.49 m | 1.35 m, 1.55 m | 1.15 d (13.1) | 1.15 d (13.1) | |||
| 22 | 1.55 m; 1.18 m | 1.55 m; 1.18 m | 2.29 m | 1.51 m, 1.37 m | 1.55 m, 1.16 m | 1.82 m, 2.04m | 1.97 m | 1.95 m | 1.46 m, 1.27 m | 1.46 m, 1.27 m | 1.83 m | 1.95 m | |
| 23 | 5.43 dt(15.0,7.0) | 5.43 dt (15.0,7.0) | 5.52 dt (11.0,7.8) | 5.53 dt(15.0,7.0) | 2.26 m, 2.16 m | 1.24 s | 1.04 s | 1.24 s | 1.04 s | 1.05 s | 0.83 s | 0.99 s | 1.22 s |
| 24 | 5.43 d (15.0) | 5.43 d (15.0) | 6.03 d (11.0) | 5.54 d (15.0) | 5.10 t (7.1) | 1.02 s | 0.82 s | 1.02 s | 0.82 s | 0.84 s | 0.75 s | 0.95 s | 0.95 s |
| 25 | - | - | - | - | - | 0.93 s | 0.75 s | 0.92 s | 0.68 s | 0.99 s | 0.85 s | 0.72 s | 0.88 s |
| 26 | 1.13 s | 1.13 s | 4.79 brs | 1.18 s | 1.40 s | 1.04 s | 0.85 s | 1.06 s | 0.85 s | 1.02 s | 0.71 s | 0.79 s | 1.04 s |
| 27 | 1.13 s | 1.13 s | 1.76 s | 1.18 s | 1.25 s | 1.30 s | 1.09 s | 1.24 s | 1.04 s | 1.18 s | 1.09 s | 1.06 s | 1.07 s |
| 28 | 0.69 s | 0.69 s | 0.79 s | 0.87 s | 0.89 s | - | - | - | - | 0.88 s | 0.93 s | 0.93 s | 0.93 s |
| 29 | 0.78 s | 0.78 s | 1.21 brs | 0.68 s | 0.97 s | 0.97 s | 0.87 s | 0.88 d | 0.90 d | 0.92 s | 0.86 s | 0.80 d (5.9) | 1.01 d (5.7) |
| 30 | 0.62 s | 0.62 s | 0.72 s | 0.62 s | 0.82 s | 1.02 s | 0.81 s | 0.97 d | 0.87 d | 0.92 s | 1.02 s | 0.87 d (7.6 ) | 0.98 d (7.4) |
| 1′ | - | - | - | 3.68 t (7.1) | - | 11.5 br s | 11.5 br s | ||||||
| 2′ | - | - | - | 1.52 m | - | ||||||||
| 3′ | - | - | - | 1.24 m | - | ||||||||
| 4′ | - | - | - | 1.26 m | - | ||||||||
| 5′ | - | - | - | 0.66 t (7.3) | - | ||||||||
Table 2: 1H (500 MHz) NMR data for compounds 1-5 in CDCl3 (δH in ppm)
Antimicrobial Activity
The n-hexane, ethyl acetate, methanol crude extracts (EAP4; EAP2 and PMRE, respectively) and isolated compounds 1 and 3 were tested for their antimicrobial activities (Tables 3-5)
| Samples | Microorganisms Tested | ||
|---|---|---|---|
| E. coli | P. aeruginosa | Salmonella Spp | |
| EAP4 | - | - | - |
| EAP2 | - | - | - |
| PMRE | - | - | - |
| 1 | - | - | 75 |
| 3 | - | - | 46 |
| Reference: Control disc with (10% DMSO) | - | - | - |
| Amoxicillin | - | 6 | - |
| Not active | |||
Table 3: In vitro antibacterial activities (diameter of the zone of inhibition-in mm) of the stock solutions of the n-hexane, ethyl acetate and methanol crude extracts and isolated compound.
| Isolated pure compounds | Microorganisms Tested | ||
|---|---|---|---|
| E. coli | Ps. aeruginosa | Salmonella spp | |
| 1 | - | - | 0.1- 0.15 |
| 3 | - | - | 0.15- 0.2 |
| Amoxicillin | - | 0.4 | - |
| - Not active | |||
Table 4:.Minimal inhibitory concentration MIC (mg/ml) of the isolated compounds showing antibacterial activities.
| Samples | Microorganisms Tested | ||
|---|---|---|---|
| S. aureus 209 | E. coli WF+ | Candida albicans 62I | |
| PMRE | 0 | 0 | 0 |
| EAP2 | 0 | 0 | 0 |
| EAP4 | 184 | 0 | 92 |
| Netilmicin | 5250 | 2400 | NT |
| 5-fluocytocine | NT | NT | 0.01 |
Table 5:.Minimal inhibitory concentration MIC (mg/ml) of the n-hexane, ethyl acetate and methanol crude extracts showing antibacterial activities
The ethyl acetate (EAP2) and methanol (PMRE) crude extracts were inactive against Escherichia coli, Pseudomonas aeruginosa, Salmonella typhi, S. aureus 209 and Candida albicans 62I (Tables 3, 5) while the lowest Minimum Inhibitory Concentration (MIC) of EAP4 was observed for S. aureus and Candida albicans (Table 5). The MIC values recorded in Table 4 were interestingly very low (0.1-0.2 mg/ml) indicating a very high antibacterial activity of compounds 1 and 3. Their antibacterial activities were found to be superior to those of Amoxicillin (0.1-0.2 versus 0.4). Their antibacterial properties against Salmonella may explain the traditional use of propolis (PROMAX-C) in the treatment of infectious diseases in Cameroon. The n-hexane crude extract (EAP4) exhibited also moderate but broad spectrum activity against both S. aureus and Candida albicans (Table 5). This confirms the fact that terpenoids contribute to the biological properties of propolis besides phenolic compounds and flavonoids [33, 34]. The specific inhibitory effects of compounds 1 and 3 against Salmonella might be due to the fact that they are pure compounds thereby lacking synergistic substances. These two cycloartane-type triterpenes may be exploited for the treatment of salmonellosis if they exhibit antimicrobial effects against other strains of non-typhoid Salmonella. On the contrary, the broad spectrum activities of nhexane crude extract (EAP4) against S. aureus and Candida albicans could explain the presence of several synergistic bioactive compounds. The strong inhibitory activities of compounds 1 and 3 against Gram negative bacteria may suggest that they could exhibit greater activities against Gram positive organisms given the fact that propolis extracts in general have been reported to be more harmful to Gram positive organisms [9, 14, 35 -36].
Therefore it seems that the presence of significant amounts of triterpenoids, combined with other bioactive compounds, is responsible for the broad spectrum of microorganisms inhibited by the n-hexane extract of propolis sample. Bankova et al. [33] found no inhibitory activity of Brazilian and Bulgarian propolis extracts against a Gram negative bacterium strain, E. coli. Netilmicin did not inhibit Candida albicans, but it inhibited S. aureus and E. coli tested. Moreover, S. aureus and E. coli were not inhibited by 5-fluocytocine but, it inhibited Candida albicans.
In general, no correlation could be established between extracts composition and their antimicrobial spectrum, since, similar antimicrobial activities were observed among samples with entirely different chemical composition. Although more than 400 constituents have been identified in propolis samples, biological activity is mainly due to a few classes of substances such as flavonoids, terpenes, phenolic acids and their esters, which have been reported to possess antimicrobial activities, and in combination considered to act synergistically [37]. This could offer an explanation for the selective and strong antimicrobial activity of propolis from different regions of Cameroon, as their extracts were very rich in terpenes and aromatic compounds (flavonoids, phenolic acids and esters). It also confirms the known ability of bees to collect the best agents to protect their hives against bacterial and fungal infections. Despite differences in the chemical composition of propolis from different geographical locations, the propolis extracts studied exhibited similar antibacterial and antifungal activities. Given the non-toxic and natural origin of propolis and the results obtained on their antimicrobial action, it is concluded that, besides their potential pharmaceutical use, low concentrations of the propolis balsams studied could be efficient protective agents for use as microbicidal additives in food systems, especially in fermented products, aiming to selectively inhibit the growth of pathogenic bacteria.
However, further pharmacological and toxicity studies currently going on in the laboratory are necessary to establish if they could be safely used as topical antimicrobial agents.
The results of our study have revealed new data about the chemical components of propolis. These results suggested that the propolis from Ngaoundal was rich in cycloartane, oleanane and ursane type triterpenoids. The plants origin of this propolis sample could be Mangifera indica (mango), Orthosiphon stamineus, Boswellia sacra, Boswellia neglecta and Melipona beecheii, Ficus exasperate, Byrosonima fagifolia, Byrosonima crassifolia, Lavandula officinalis, Euphorbia dendroides, Pachysandra terminalis, Melandrium firmum.
Further purification of the propolis fractions will be done with the aim of obtaining more constituents and their biological activities such as anticonvulsant and antiviral, will be researched in our laboratory using several techniques. We do hope, this work will attract the attention of scientists to further research on propolis. The results of this Cameroonian propolis on the tested organisms in this study are encouraging in comparison with the previous works [36]. It is interesting to note that compounds 1 and 3 are more active than the original extract (EAP4) but lose their broad spectrum potency in the course of purification. However, further pharmacological studies currently going on in the laboratory are necessary to establish if they could be safely used as topical antimicrobial agents.
We are grateful to the members of AHIMSA (Non-Governmental Organization, in Spain) for supporting part of this work through a financial help to Sakava. P.; S.L. and L.V.E thank the ARC (research contract AUWB-2010—10/15UMONS-5), the FNRS, ENCITE program, the COST TD1004 (Theranostics imaging and therapy: An action to develop novel nanosized systems for imaging-guided drug delivery), the UIAP VII program, the European Network of Excellence EMIL (European Molecular Imaging Laboratories) program LSCH-2004503569 and the Center for Microscopy and Molecular Imaging (CMMI, supported by the European Regional Development Fund and the Walloon Region).
Conceptualization, S.P.; T.E.; J.T.M and T.F.M.; Validation, T.E.; J.T.M. and A.D.T.A.; Software, S.L.; C.H.; V.E.L. and S.P.; Data curation and Formal analysis, S.L.; C.H.; V.E.L. and S.P.; Investigation and Methodology, S.P. and T.F.M.; Supervision, T.E.; J.T.M.; A.D.T.A.; S.L.; C.H. and V.E.L. Writing – Original Draft, S.P.; review and editing, T.E.; T.F.M.; J.T.M. and A.D.T.A.; Visualization, S.P.; T.E.; J.T.M.; T.F.M.; A.D.T.A.; S.L.; C.H.; V.E.L. All authors read and approved the final manuscript.
The authors declare no conflict of interest.
Citation: Paul S (2021) Two New Cycloartenol and Some Pentacyclic Triterpenoids from Cameroonian Propolis (Ngaoundal, AD Region) and Evaluation of their Antimicrobial Activity, Nat Prod Chem Res. 9:415.
Published: 02-Sep-2021
Copyright: © 2021 Paul S. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.